Congratulations to CAI HDR students Joshua Harbort and Tom Shaw whose publications were selected as joint winners of Round 2 of the CAI 2020 HDR publication competition.
Harbort, Joshua S; Podgorski, Matthew N; Coleman, Tom; Stok, Jeanette E; Yorke, Jake A; Wong, Luet-Lok; Bruning, John B; Bernhardt, Paul V; De Voss, James J; Harmer, Jeffrey R; Bell, Stephen G; "Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases" Biochemistry, 2020, Vol.59(9), doi: 10.1021/acs.biochem.0c00027
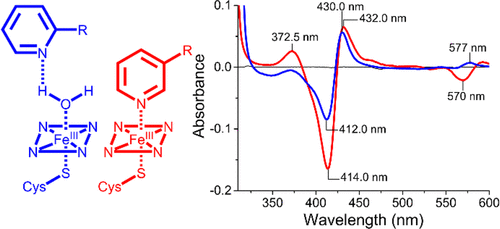 ABSTRACT: The cytochrome P450 superfamily of heme monooxygenases catalyzes important chemical reactions across nature. The changes in the optical spectra of these enzymes, induced by the addition of substrates or inhibitors, are critical for assessing how these molecules bind to the P450, enhancing or inhibiting the catalytic cycle. Here we use the bacterial CYP199A4 enzyme (Uniprot entry Q2IUO2), from Rhodopseudomonas palustris HaA2, and a range of substituted benzoic acids to investigate different binding modes. 4-Methoxybenzoic acid elicits an archetypal type I spectral response due to a ≥95% switch from the low- to high-spin state with concomitant dissociation of the sixth aqua ligand. 4- (Pyridin-3-yl)- and 4-(pyridin-2-yl)benzoic acid induced different type II ultraviolet−visible (UV−vis) spectral responses in CYP199A4. The former induced a greater red shift in the Soret wavelength (424 nm vs 422 nm) along with a larger overall absorbance change and other differences in the α-, β-, and δ-bands. There were also variations in the ferrous UV−vis spectra of these two substrate-bound forms with a spectrum indicative of Fe−N bond formation with 4-(pyridin-3-yl)benzoic acid. The crystal structures of CYP199A4, with the pyridinyl compounds bound, revealed that while the nitrogen of 4-(pyridin-3-yl)benzoic acid is coordinated to the heme, with 4-(pyridin-2-yl)benzoic acid an aqua ligand remains. Continuous wave and pulse electron paramagnetic resonance data in frozen solution revealed that the substrates are bound in the active site in a form consistent with the crystal structures. The redox potential of each CYP199A4−substrate combination was measured, allowing correlation among binding modes, spectroscopic properties, and the observed biochemical activity.
ABSTRACT: The cytochrome P450 superfamily of heme monooxygenases catalyzes important chemical reactions across nature. The changes in the optical spectra of these enzymes, induced by the addition of substrates or inhibitors, are critical for assessing how these molecules bind to the P450, enhancing or inhibiting the catalytic cycle. Here we use the bacterial CYP199A4 enzyme (Uniprot entry Q2IUO2), from Rhodopseudomonas palustris HaA2, and a range of substituted benzoic acids to investigate different binding modes. 4-Methoxybenzoic acid elicits an archetypal type I spectral response due to a ≥95% switch from the low- to high-spin state with concomitant dissociation of the sixth aqua ligand. 4- (Pyridin-3-yl)- and 4-(pyridin-2-yl)benzoic acid induced different type II ultraviolet−visible (UV−vis) spectral responses in CYP199A4. The former induced a greater red shift in the Soret wavelength (424 nm vs 422 nm) along with a larger overall absorbance change and other differences in the α-, β-, and δ-bands. There were also variations in the ferrous UV−vis spectra of these two substrate-bound forms with a spectrum indicative of Fe−N bond formation with 4-(pyridin-3-yl)benzoic acid. The crystal structures of CYP199A4, with the pyridinyl compounds bound, revealed that while the nitrogen of 4-(pyridin-3-yl)benzoic acid is coordinated to the heme, with 4-(pyridin-2-yl)benzoic acid an aqua ligand remains. Continuous wave and pulse electron paramagnetic resonance data in frozen solution revealed that the substrates are bound in the active site in a form consistent with the crystal structures. The redox potential of each CYP199A4−substrate combination was measured, allowing correlation among binding modes, spectroscopic properties, and the observed biochemical activity.
Shaw, Thomas ; York, Ashley; Ziaei, Maryam; Barth, Markus; Bollmann, Steffen; "Longitudinal Automatic Segmentation of Hippocampal Subfields (LASHiS) using multi-contrast MRI" NeuroImage, 2020, Vol.218, doi: doi.org/10.1016/j.neuroimage.2020.116798
ABSTRACT: The volumetric and morphometric examination of hippocampus formation subfields in a longitudinal manner using in vivo MRI could lead to more sensitive biomarkers for neuropsychiatric disorders and diseases including Alzheimer’s disease, as the anatomical subregions are functionally specialised. Longitudinal processing allows for increased sensitivity due to reduced confounds of inter-subject variability and higher effect-sensitivity than cross-sectional designs. We examined the performance of a new longitudinal pipeline (Longitudinal Automatic Segmentation of Hippocampus Subfields [LASHiS]) against three freely available, published approaches. LASHiS automatically segments hippocampus formation subfields by propagating labels from cross-sectionally labelled time point scans using joint-label fusion to a non-linearly realigned ‘single subject template’, where image segmentation occurs free of bias to any individual time point. Our pipeline measures tissue characteristics available in in vivo high-resolution MRI scans, at both clinical (3 T) and ultra-high field strength (7 T) and differs from previous longitudinal segmentation pipelines in that it leverages multi-contrast information in the segmentation process. LASHiS produces robust and reliable automatic multi-contrast segmentations of hippocampus formation subfields, as measured by higher volume similarity coefficients and Dice coefficients for test-retest reliability and robust longitudinal Bayesian Linear Mixed Effects results at 7 T, while showing sound results at 3 T. All code for this project including the automatic pipeline is available at https://github.com/CAIsr/LASHiS.



